APPLICATION OF TECHNOLOGY:
- Comparative genomics
ABSTRACT:
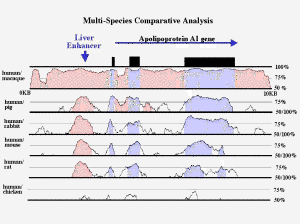
VISualization Tool for Alignments is a program to depict long sequence alignments of DNA from two or more organisms with various types of annotation in a clear and easily interpretable format. Originally it was developed to locate conserved sequences in syntenic regions.
Key Features:
- Clean graphical output, allowing for easy identification of sequence similarities and differences
- Easily configurable, enabling the visualization of alignments of up to several megabases at different levels of resolution
- Displays alignments of draft sequences
- Displays sequence annotations such as repeats, coding exons, UTRs and more
The VISTA plot is based on moving a user-specified window over the entire alignment and calculating the percent identity over the window at each base pair.
AVID is an alignment program designed to work seamlessly with VISTA. AVID globally aligns DNA sequences of arbitrary length for the purpose of annotation and biological discovery using syntenic genomic sequences from two organisms.
Key Features:
- Aligns hundreds of kilobases quickly
- Highly accurate and able to detect weak homologies
- Able to handle one sequence in draft by ordering and orienting the contigs automatically
- Fast alignment of similar sequences is useful for alignments of primate sequences or comparison of assemblie
AVID works by recursively finding strong anchors from the collection of maximal matches in the sequences.
STATUS: To purchase a license for VISTA or AVID or to learn more go to: http://www-gsd.lbl.gov/vista/
FOR MORE INFORMATION PLEASE SEE:
REFERENCE NUMBER: CR-1690; CR-1754