APPLICATIONS OF TECHNOLOGY:
- Determines three-dimensional (3D) macromolecular structure using crystallography (X-ray, neutron, or electron) or cryo-electron microscopy data.
BENEFITS:
- Provides a comprehensive, highly-automated software package for determining macromolecular structure.
- Increases the throughput of structure solutions.
- Designed for users at all levels, from beginners to experts. Users can easily access the programs through the Phenix graphical user interface (GUI) and/or command line.
- Focuses on rapid development and bug fixing.
- Provides integration with molecular viewers (Coot and Pymol; coming: ChimeraX).
- Offers convenient ligand handling tools (automated ligand fitting and restraints builders).
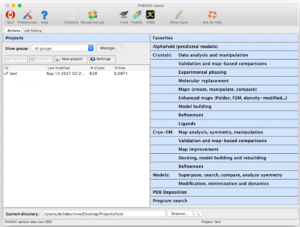
Phenix GUI showing the project and task management on the left, and the list of program categories on the right.
BACKGROUND:
Knowledge of molecular structures is critical to understanding biological processes and to developing new therapeutics against diseases. Crystallography (X-ray, neutron, and electron) and cryo-electron microscopy are powerful methods for determining 3D macromolecular structures. The overall structure-solution workflow is similar for these techniques, but nuances exist because their reduced experimental data have different properties. Phenix is a comprehensive software package for macromolecular structure determination that handles data acquired using any of these techniques.
TECHNOLOGY OVERVIEW:
Researchers at Lawrence Berkeley National Lab and other institutions have developed a comprehensive software package, called Phenix (Python-based Hierarchical ENvironment for Integrated Xtallography), that efficiently and accurately determines macromolecular structures from crystallography and cryo-electron microscopy data.
The Phenix package performs all of the steps required to determine a macromolecular structure. For X-ray diffraction data, these include assessing the data quality, experimental phasing, molecular replacement, building an atomic model, refining and validating the model, and preparing structure deposition into the World-Wide Protein Data Bank. For cryo-electron microscopy data, these include assessing map quality, improving the map, building, refining and validating a model, and preparing structure deposition.
The programs developed for each of these steps are highly automated. Thus, Phenix provides rapid structure solutions for both X-ray crystallography and cryo-electron microscopy data—ultimately enabling increased throughput.
The Phenix programs are run through a user-friendly interface and/or by command line. The extensive online manual describes GUI and command-line versions of individual programs and includes overviews, tutorials, and FAQs. The Phenix Tutorials YouTube channel provides introductory tutorial videos. Each program sets defaults based on their most common use, but also allows changing the defaults to better meet users’ needs.
Phenix licensees must join the Phenix Consortium; we are currently seeking industrial partners to join the consortium to further the development of the Phenix software. For more information, please visit https://phenix-online.org/consortium.
FOR MORE INFORMATION:
CONTRIBUTORS:
- Lawrence Berkeley National Laboratory (Paul Adams group)
- New Mexico Consortium and Los Alamos National Laboratory (Tom Terwilliger’s group)
- Cambridge University (Randy Read’s group)
- Duke University (the Richardsons’ group)
- UTHealth (Matt Baker’s group)
STATUS: Copyrighted.
OPPORTUNITIES: Available for licensing (free for academia, fee for commercial use).